Cornell University
Musharoff Research Lab
Current Research Areas
Our lab emphasizes integrating data from both human and non-human populations to understand the mechanisms behind genomic variation and its implications for health disparities. We focus on factors such as admixture, gene-environment interactions, and demographic structures that influence disease risk. Utilizing statistical and computational methods, we investigate complex traits and develop techniques for inferring demographic histories and analyzing deleterious variation. Additionally, we aim to promote the equitable inclusion of underrepresented populations in genomics, thereby enriching our insights into genetic diversity and trait architecture.
Genetic Variation and Disease Risk in Diverse Populations
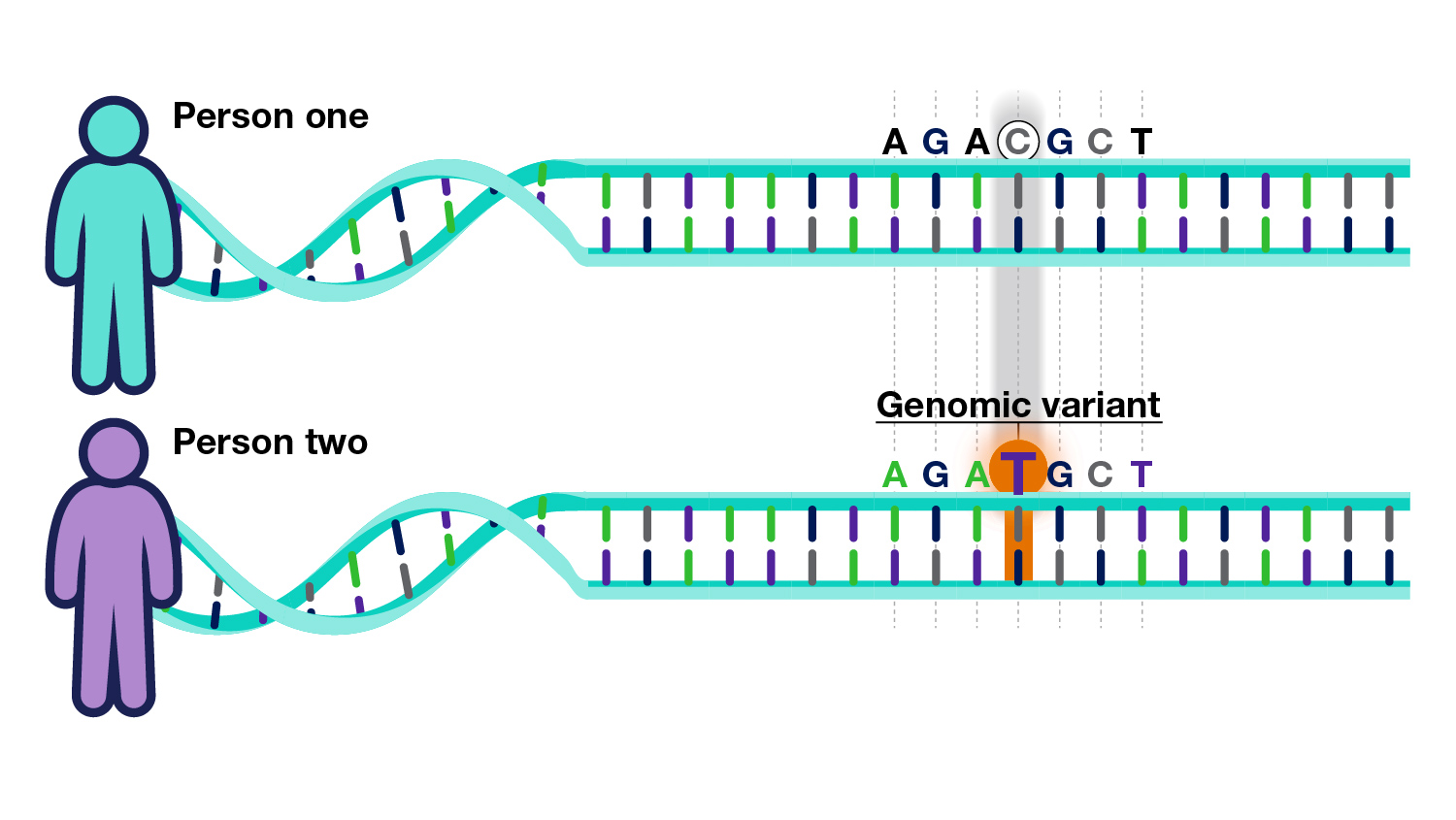
Current research efforts in defining genetic variation and disease risk in diverse populations focus on addressing the underrepresentation of non-European ancestries in genomic studies. The limitations of existing polygenic risk scores (PRSs) have highlighted the need for tailored models that consider genetic diversity. By leveraging large-scale Genome-Wide Association Studies (GWASs) and integrating data from diverse populations, researchers aim to refine risk assessments for complex diseases, thereby enhancing the applicability of genetic findings across different demographic groups. To further enhance understanding, studies are increasingly incorporating Electronic Health Records (EHRs) to link genetic data with clinical outcomes, allowing for a comprehensive evaluation of genetic risk factors in real-world settings. Machine learning algorithms for model refinement are being employed to improve predictive accuracy. By fostering inclusivity in genetic research, these efforts aim to develop more effective precision medicine strategies, ultimately addressing health disparities and promoting equitable healthcare for diverse populations.
Population and Statistical Genetics
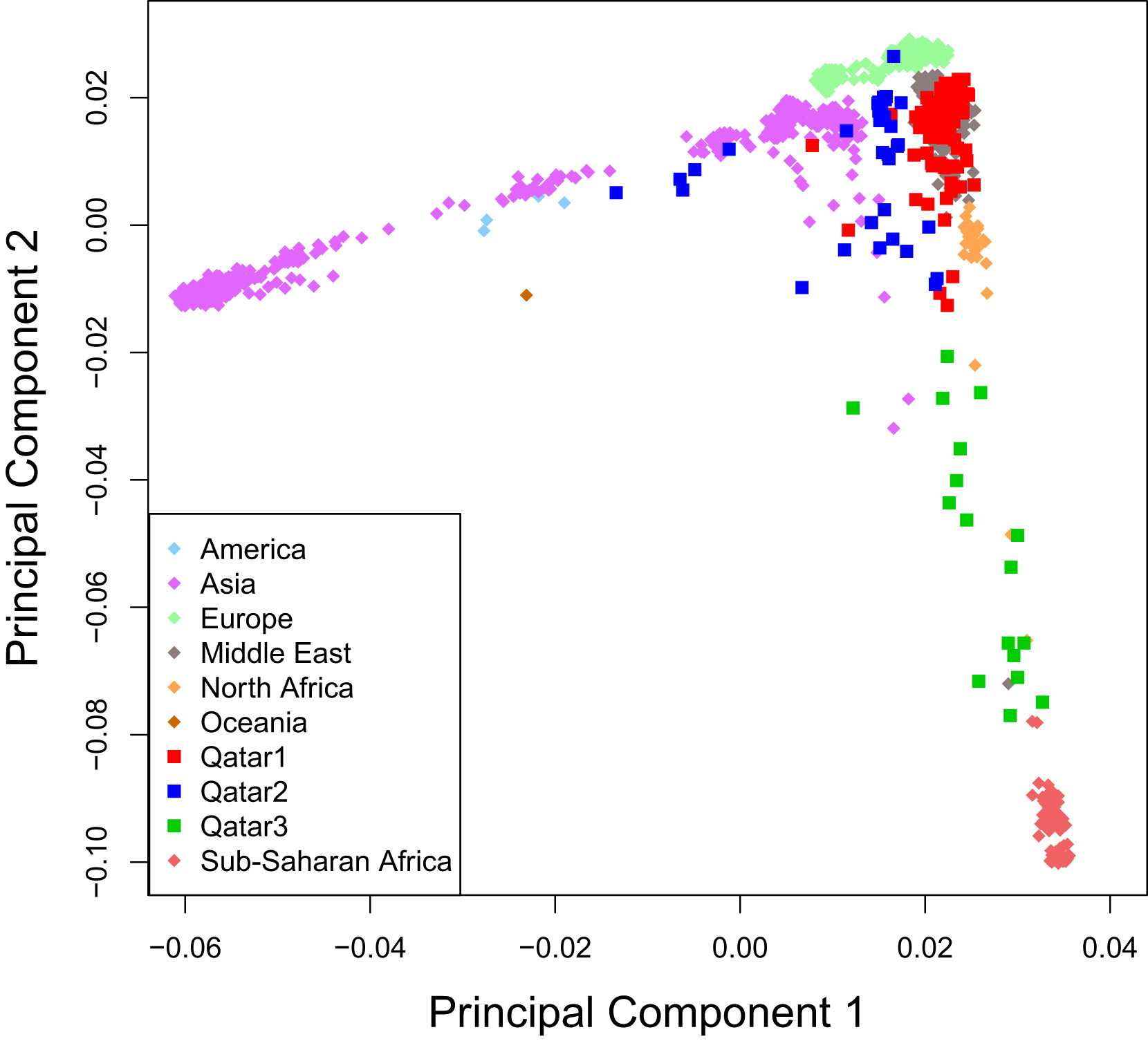
Research in population genetics and statistical genetics has advanced significantly, focusing on methods to understand population structure and genetic diversity. Genetic data collected from millions of individuals across various populations is being utilized to broaden our understanding of genetic relationships within and among populations. Notable developments include the Structure software for model-based clustering, Principal Component Analysis (PCA) to reveal geographic patterns, and the use of coalescent theory and Hidden Markov Models to estimate recombination rates and ancestry patterns. Projects in the Musharoff lab address population structure, demographic influences, and the interplay of selection and history, facilitating insights into human evolutionary dynamics.
Gene-Environment Interactions
Gene-environment (GxE) research delves into the intricate interactions between genetic and environmental factors that influence traits. Rather than viewing traits as simple additive combinations of genetic and environmental components, this field acknowledges that complex relationships define them. Gene-gene interactions (GxG), including epistasis, complicate the understanding of trait variance, while GxE interactions reveal how specific environmental contexts can either amplify or mitigate genetic effects. Moreover, gene-environment correlations (rGE) illustrate how genetic predispositions can shape an individual's environmental exposures, leading to feedback loops that affect traits across generations. Ongoing research in the Musharoff lab focuses on refining models to better capture these dynamic interactions, aiming to enhance our understanding of the multifaceted nature of traits and their development.